Abstract
Background:
Clinical features have been previously associated with the development of chronic immune thrombocytopenia (ITP) in children, but biomarkers to distinguish acute, self-resolving ITP from chronic disease remain elusive. Identifying biomarkers specific for chronic ITP would guide treatment decisions and also inform disease pathogenesis, which remains poorly understood. Prior studies have shown that inflammatory cytokines are elevated in chronic ITP patients as compared to acute patients, and DNA and RNA microarray data stratified chronic and acute ITP patients.
This study leverages RNA sequencing data to characterize acute and chronic ITP cohorts. RNA sequencing has the advantage of examining RNA expression differences in a systematic and unbiased manner, as opposed to prior studies which were limited by candidate approach or by array technology.
Methods:
We collected biologic materials for sequencing and clinical data from 274 pediatric ITP Texas Children's Hospital patients from July 2015 to July 2018 under an approved IRB protocol. Patients were followed prospectively and classified as (1) chronic with disease persisting >12 months or (2) acute with spontaneous resolution in <12 months. Whole blood samples were collected at two time points over disease course. For acute patients, the initial sample was collected within 1 month of initial diagnosis and the second sample was drawn at the time of disease resolution. Two samples, 3-6 months apart, were taken from known chronic ITP patients with active disease at both time points. Acute patients were excluded if they had received any ITP directed therapy. Chronic patients were excluded if they had any therapy within the six months prior to the first sample or had ever received rituximab.
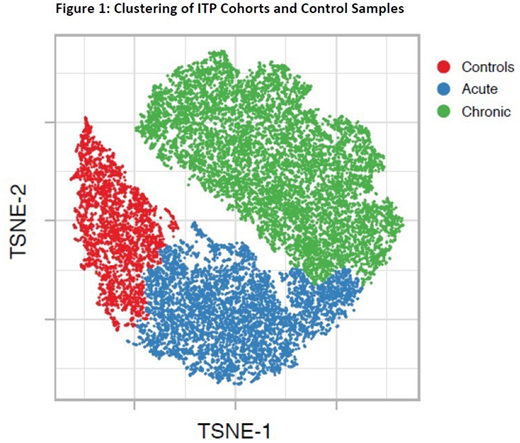
Peripheral blood mononuclear cells were isolated using a ficoll separation from whole blood, and total RNA was extracted and purified using an RNeasy Mini Kit (QIAGEN). Those sample pairs which met standards for amount and purity of RNA for both samples were sequenced. RNA-Seq libraries were generated using Ovation® Universal RNA-Seq System (NuGen), sequenced on an Illumina NextSeq 500 instrument, and generated 40 million paired end reads. In total, 9 acute pairs, 12 chronic pairs and 9 healthy controls were sequenced. All protein coding transcripts were collapsed by gene, then rank statistics were applied and scaled by z-score. Cohorts were refined such that samples met uniform quality standards. Cluster analysis was performed using t-distributed stochastic neighbor embedding (t-SNE), a nonlinear method to reduce dimensionality and identify similar structures between samples.
Results:
Over 20,000 RNA transcripts were identified in approximately 2,000 genes. This preliminary analysis focused on initial samples only, when all ITP patients had active disease. T-SNE analysis showed that healthy controls, acute, and chronic ITP patients stratify into three separate clusters (Figure 1). A hierarchical clustering heatmap demonstrated a stratification between samples from acute patients at presentation, samples from chronic patients, and samples from healthy pediatric controls. Transcripts which stratified patients via hierarchical clustering were analyzed using Gene Set Enrichment Analysis (GSEA) to provide functional annotations. Applying transcripts with high expression in active acute ITP, mid-range expression in chronic ITP, and low expression in healthy controls identified pathways which are also important in other autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, and inflammatory bowel disease. Individual transcript expression profiles were not specifically compared in this analysis, but transcripts with immune function including Fc-Gamma Receptor IIIb, CXCL8, BMP7, DEFA1, DEFA1B, HLA-DRB4, and HLA-DRB5 were differentially expressed between ITP patient samples and controls.
Conclusions:
This study is the first to use RNA sequencing to show that active, acute ITP is distinct from chronic ITP. This study confirmed that ITP patient RNA expression patterns differ from that of healthy controls. Both transcripts with differential expression between cohorts and functional annotation pathways identified patterns with relevance to autoimmunity.
Grace:Agios Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees; Agios Pharmaceuticals: Research Funding; Agios Pharmaceuticals: Consultancy. Lambert:Summus: Consultancy; Shionogi: Consultancy; Amgen: Membership on an entity's Board of Directors or advisory committees; Educational Concepts in Medicine: Consultancy; Rigel: Consultancy; Sysmex: Consultancy; CSL: Consultancy; Novartis: Membership on an entity's Board of Directors or advisory committees; Bayer: Membership on an entity's Board of Directors or advisory committees. Despotovic:Sanofi: Consultancy; Novartis: Research Funding; AmGen: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal